CytoGPS is a software tool to parse semi-structured ISCN-based karyotypes in text formats.
Validation
Built using ANTLR (ANother Tool for Language Recognition), first filters out unacceptable karyotype input. It then uses several ad-hoc algorithms to validate each cytogenetic aberration, including derivative chromosomes. Detailed error messages are displayed to the user.
ISCN Correction
For fixable parsing errors, e.g. incorrect types of brackets for cell numbers, a clean suggested revised karyotype will be presented to the user. For validation errors, a suggestion, whenever possible, will be given.
Analysis
Extracts biologically important data from text-based karyotypes, including but not limited to the number of occurrences of the biological events (loss, gain, and fusion) occurring at all the bands (or subbands) in the chromosome. This transformation makes quantitative analyses feasible which were previously not possible.
Q: Can CytoGPS software parse karyograms directly?
A: No. Unlike commercial software programs which provide karyotyping solutions following the capture of digital images, CytoGPS aims to parse semi-structured ISCN-based karyotypes in text formats. To our knowledge, there is no commercial software available to read text-based karyotypes: CytoGPS is filling in this gap. We expect the user to enter text-based karyotypes following the International System for Human Cytogenomic Nomenclature (ISCN).
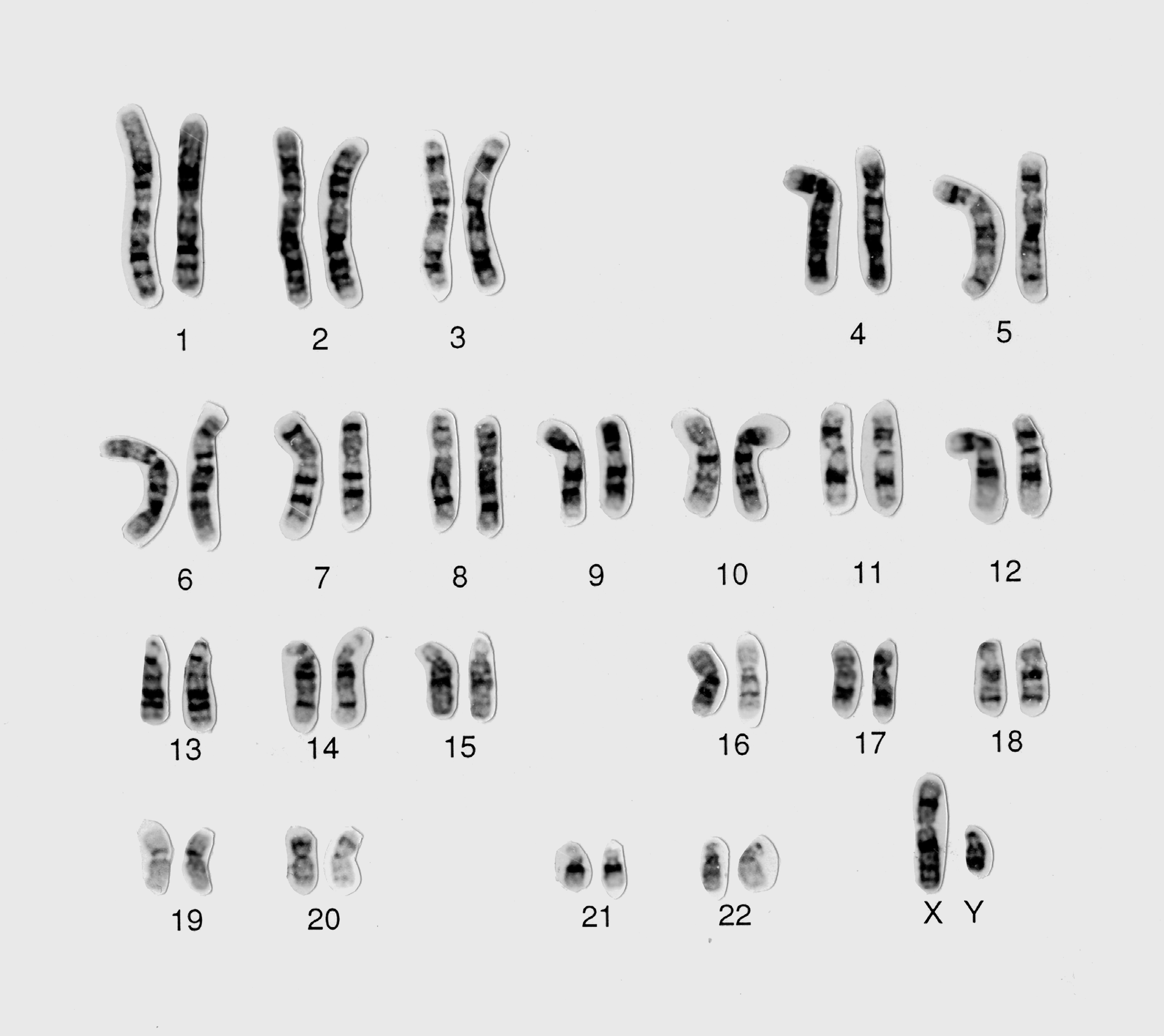
This figure is a derivative of Normal male 46,XY human karyotype by Wessex Reg. Genetics Centre, used under CC-BY (v4.0). https://wellcomecollection.org/works/zd7rdetc
Q: How can cytogeneticists and biomedical data scientists use CytoGPS software?
A: CytoGPS extracts biologically important, quantitative information for biomedical data scientists to do further analysis. These quantitative data include a tally of the cytogenetic events (loss, gain, and fusion) occurring at all G-850 bands (or subbands) in the chromosome. Other useful information provided to cytogeneticists includes number of cells in a clone, relationships among multiple clones, and detailed systems for characterizing derivative chromosomes.
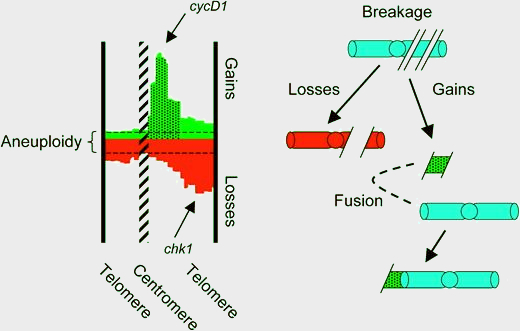
This figure is a derivative of Figure 1 from Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas by Carlos Martínez-A and Karel H.M. van Wely, used under CC BY-NC (v2.5). https://doi.org/10.1093/carcin/bgr069
Q: Can CytoGPS software parse karyotypes using high resolution techniques?
A: Not yet. Techniques utilizing fluorescent in situ hybridization (FISH) and chromosomal microarray (CMA) have become an adjunct to traditional chromosome analysis. FISH allows researchers to locate the positions of specific DNA sequences on chromosomes, while CMA detects copy number variations (CNVs) in the genomes. As major advances in human cytogenetics, both FISH and CMA have been increasingly used in karyotype reports. In the near future, we will extend CytoGPS to enable parsing of karyotypes using these high resolution techniques.
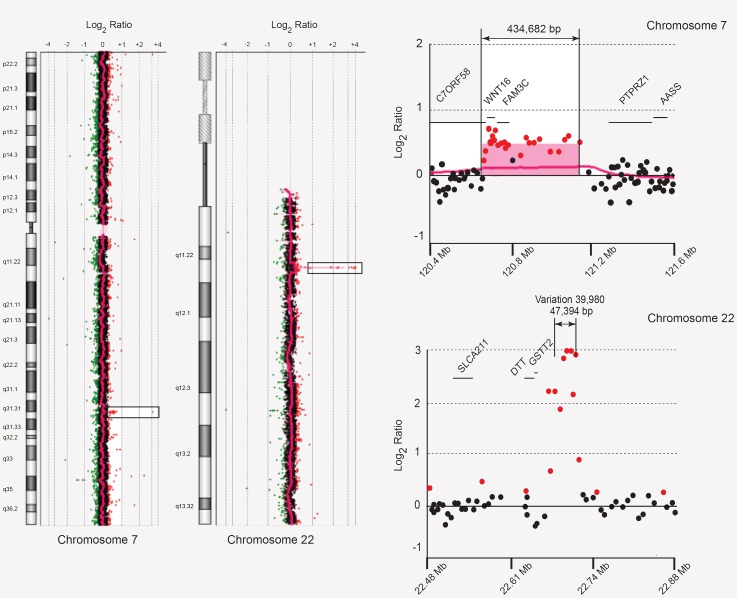
This figure is a derivative of Figure 2 from Duplication of C7orf58, WNT16 and FAM3C in an Obese Female with a t(7;22)(q32.1;q11.2) Chromosomal Translocation and Clinical Features Resembling Coffin-Siris Syndrome by Jun Zhu, Jun Qiu, Gregg Magrane, et al, used under CC-BY (v4.0). https://doi.org/10.1371/journal.pone.0052353
